ANGSD: Analysis of next generation Sequencing Data
Latest tar.gz version is (0.938/0.939 on github), see Change_log for changes, and download it here.
Safv3: Difference between revisions
| Line 21: | Line 21: | ||
##comparison | ##comparison | ||
a<- | a<-scan("newceu.saf.idx.chr1.ml") | ||
b<- | b<-as.numeric(read.table("oldceu.saf.ml")[1,]) | ||
a-b | a-b | ||
[1] 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 | [1] 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 | ||
Revision as of 17:45, 5 February 2016
We decided to update the native simple binary double format to a much more intelligent format that allows for random access. The format is described in doc/formats.pdf.
This page will contain the impact of this new format in downstream analysis.
One population analysis
#old master
angsd version: 0.801-27-ga699b44 (htslib: 1.2.1-62-g35746af) build(May 5 2015 03:38:17)
#new new saf
angsd version: 0.801-54-gcf1a12d-dirty (htslib: 1.2.1-62-g35746af) build(May 6 2015 23:34:27)
##old
../master/angsd -anc hg19ancNoChr.fa -dosaf 1 -b /space/genomes/1000g/lowC2014/filelists/ceu.ricco.list -gl 1 -P 5 -out oldceu -rf rf
../master/misc/realSFS oldceu.saf 36 -nSites 213376207 -P 20 >oldceu.saf.ml
##new
../angsd/angsd -anc hg19ancNoChr.fa -dosaf 1 -b /space/genomes/1000g/lowC2014/filelists/ceu.ricco.list -gl 1 -P 5 -out newceu -rf rf
../angsd/misc/realSFS ../nsfs/newceu.saf.idx -P 16 -r 1 >ceu.chr1
##comparison
a<-scan("newceu.saf.idx.chr1.ml")
b<-as.numeric(read.table("oldceu.saf.ml")[1,])
a-b
[1] 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
[6] 0.000000e+00 0.000000e+00 -1.248518e-10 4.059244e-10 -3.843052e-10
[11] 4.952888e-10 -2.465176e-10 7.169737e-11 0.000000e+00 0.000000e+00
[16] 0.000000e+00 0.000000e+00 -4.288667e-11 0.000000e+00 0.000000e+00
[21] 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
[26] 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
[31] 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00
[36] 0.000000e+00 0.000000e+00
range(a-b)
[1] -3.843052e-10 4.952888e-10
barplot(rbind(a,b)[,-c(1,37)],be=T,legend=c("new","old"),col=1:2)
Two population analysis
##old(master) -> angsd version: 0.801-28-gbab908a (htslib: 1.2.1-62-g35746af) build(May 9 2015 14:50:33) ##new(newsaf)-> angsd version: 0.801-61-g48f06d8-dirty (htslib: 1.2.1-62-g35746af) build(May 9 2015 08:49:40) ##old version required a run for each population, to find the intersect and then limit the analysis to the intersect. ##Here are all 4 commands ==> oldceu2.arg <== ../master/angsd -anc hg19ancNoChr.fa -dosaf 1 -b /space/genomes/1000g/lowC2014/filelists/ceu.ricco.list -gl 1 -P 5 -out oldceu2 -r 1 -sites intersect.txt ==> oldceu.arg <== ../angsd/angsd -anc hg19ancNoChr.fa -dosaf 1 -b /space/genomes/1000g/lowC2014/filelists/ceu.ricco.list -gl 1 -P 5 -out oldceu -r 1 ==> oldyri2.arg <== ../master/angsd -anc hg19ancNoChr.fa -dosaf 1 -b /space/genomes/1000g/lowC2014/filelists/yri.ricco.list -gl 1 -P 5 -out oldyri2 -r 1 -sites intersect.txt ==> oldyri.arg <== ../angsd/angsd -anc hg19ancNoChr.fa -dosaf 1 -b /space/genomes/1000g/lowC2014/filelists/yri.ricco.list -gl 1 -P 5 -out oldyri -r 1 ##with intersect found like gunzip -c oldceu.saf.pos.gz oldyri.saf.pos.gz|sort -S 50%|uniq -d|sort -k1,1 -S 50% >intersect.txt ##The old saf files are very big so we had to limit the analysis to 100mio sites ../master/misc/realSFS 2dsfs oldceu2.saf oldyri2.saf 36 52 -nSites 100000000 -P 20 >oldceu2.oldyri2.ml.100mb ##the new format is much simpler here we simply did ==> newceu.arg <== ../angsd/angsd -anc hg19ancNoChr.fa -dosaf 1 -b /space/genomes/1000g/lowC2014/filelists/ceu.ricco.list -gl 1 -P 5 -out newceu -r 1 ==> newyri.arg <== ../angsd/angsd -anc hg19ancNoChr.fa -dosaf 1 -b /space/genomes/1000g/lowC2014/filelists/yri.ricco.list -gl 1 -P 5 -out newyri -r 1 ../angsd/misc/realSFS ../nsfs/newceu.saf.idx ../nsfs/newyri.saf.idx -P 32 -r 1 -nSites 100000000 >ceu.yri.chr1.100mb
and comparing in R
norm<-function(X) X/sum(X)
a<-exp(read.table("oldceu2.oldyri2.100mb",nrow=37))
b<-norm(scan("newceu.newyri.chr1.100mb"))
b<-matrix(b,ncol=53,byrow=T)
> range(a-b)
[1] -1.724603e-07 2.850303e-09
> png("safv3to.png")
> barplot(rbind(colSums(a),colSums(b))[,-c(1,53)],be=T,main="marginal 2dsfs YRI",legend=c("safv1","safv3"),col=1:2)
> dev.off()
X11cairo
2
> png("safv3en.png")
> barplot(rbind(rowSums(a),rowSums(b))[,-c(1,37)],be=T,main="marginal 2dsfs CEU",legend=c("safv1","safv3"),col=1:2)
> dev.off()
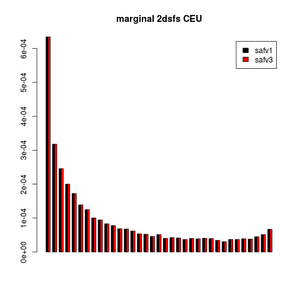
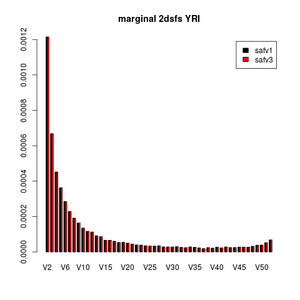
This looks very nice, right?
2 population analysis with simulated data
After doing this analysis I noticed that I had put a zero too much on the -reglen option for the simulated data with invariable sites. And still the method works fine. The result is that we still find the sfs nicely eventhough we only have 1/10 of the variability that we would expect from human data. So this is very good.
##angsd new: ->-> angsd version: 0.801-51-g156039a (htslib: 1.2.1-69-gb79f40a) build(May 7 2015 15:31:53)
##angsd old: -> angsd version: 0.801-27-ga699b44 (htslib: 1.2.1-69-gb79f40a) build(May 7 2015 15:30:06)
norm <- function(x) x/sum(x)
if(FALSE){
##generate data DONT RUN
if(FALSE){
##simulate data with msms
nRep <- 10
nPop1 <- 24
nPop2 <- 16
cmd <- paste("msms -ms",nPop1+nPop2,nRep,"-t 930 -r 400 -I 2",nPop1,nPop2,"0 -g 1 9.70406 -n 1 2 -n 2 1 -ma x 0.0 0.0 x -ej 0.07142857 2 1 >msoutput.txt ",sep=" ")
system(cmd)
##system("msms -ms 40 1 -t 930 -r 400 -I 2 20 20 0 -g 1 9.70406 -n 1 2 -n 2 1 -ma x 0.0 0.0 x -ej 0.07142857 2 1 >msoutput.txt ")
source("readms.output.R")
}
if(FALSE){
##use R to calculate SFS for each pop and 2dsfs
source("../R/readms.output.R")
a<- read.ms.output(file="msoutput.txt")
p1.d <- unlist((sapply(a$gam,function(x) colSums(x[1:nPop1,]))))
p2.d <- unlist((sapply(a$gam,function(x) colSums(x[-c(1:nPop1),]))))
par(mfrow=c(1,2))
barplot(table(p1.d))
barplot(table(p2.d))
sfs.2d <- sapply(0:nPop1,function(x) table(factor(p2.d[p1.d==x],levels=0:nPop2)))
}
if(FALSE){
##generate ANGSD inputfiles without invariable sites and run it
system("../misc/msToGlf -in msoutput.txt -out raw -singleOut 1 -regLen 0 -depth 8 -err 0.005")
system("../misc/splitgl raw.glf.gz 20 1 12 >pop1.glf.gz")
system("../misc/splitgl raw.glf.gz 20 13 20 >pop2.glf.gz")
system("echo \"1 250000000\" >fai.fai")
system("../angsd -glf pop1.glf.gz -nind 12 -doSaf 1 -out pop1 -fai fai.fai -issim 1")
system("../angsd -glf pop2.glf.gz -nind 8 -doSaf 1 -out pop2 -fai fai.fai -issim 1")
system("../misc/realSFS pop1.saf.idx >pop1.saf.idx.ml")
system("../misc/realSFS pop2.saf.idx >pop2.saf.idx.ml")
system("../misc/realSFS pop1.saf.idx pop2.saf.idx >pop1.pop2.saf.idx.ml")
}
if(FALSE){
pop1 <- exp(scan("pop1.saf.idx.ml"))
pop2 <- exp(scan("pop2.saf.idx.ml"))
pop1.pop2 <- matrix(exp(scan("pop1.pop2.saf.idx.ml")),nPop1+1,byrow=T)
par(mfrow=c(1,2))
barplot(rbind(norm(table(p1.d)),pop1),be=T,main="only varsites pop1")
barplot(rbind(norm(table(p2.d)),pop2),be=T,main="only varsites pop2")
range(norm(sfs.2d)-t(pop1.pop2))
##[1] -0.0005150658 0.0004685074
barplot(rbind(rowSums(norm(t(sfs.2d))),rowSums(pop1.pop2)),be=T)
barplot(rbind(colSums(norm(t(sfs.2d))),colSums(pop1.pop2)),be=T)
}
if(FALSE){
##simulate angsd inputfiles with invariable sites and run it
system("../misc/msToGlf -in msoutput.txt -out raw -singleOut 1 -regLen 10000000 -depth 8 -err 0.005")
system("../misc/splitgl raw.glf.gz 20 1 12 >pop1.glf.gz")
system("../misc/splitgl raw.glf.gz 20 13 20 >pop2.glf.gz")
system("echo \"1 250000000\" >fai.fai")
system("../angsd -glf pop1.glf.gz -nind 12 -doSaf 1 -out pop1 -fai fai.fai -issim 1")
system("../angsd -glf pop2.glf.gz -nind 8 -doSaf 1 -out pop2 -fai fai.fai -issim 1")
system("../misc/realSFS pop1.saf.idx >pop1.saf.idx.ml")
system("../misc/realSFS pop2.saf.idx >pop2.saf.idx.ml")
system("../misc/realSFS pop1.saf.idx pop2.saf.idx >pop1.pop2.saf.idx.ml")
}
if(FALSE){
pop1 <- norm(exp(scan("pop1.saf.idx.ml"))[-1])
pop2 <- norm(exp(scan("pop2.saf.idx.ml"))[-1])
pop1.pop2 <- matrix(exp(scan("pop1.pop2.saf.idx.ml")),nPop1+1,byrow=T)
par(mfrow=c(1,2))
barplot(rbind(norm(table(p1.d)[-1]),pop1),be=T,main="varsites pop1")
barplot(rbind(norm(table(p2.d)[-1]),pop2),be=T,main="varsites pop2")
pop1.pop2[1,1] <- 0
pop1.pop2[nrow(pop1.pop2),ncol(pop1.pop2)] <- 0
pop1.pop2 <- norm(pop1.pop2)
range(norm(sfs.2d)-t(pop1.pop2))
#[1] -0.0005047007 0.0007018686
barplot(rbind(rowSums(norm(t(sfs.2d))),rowSums(pop1.pop2)),be=T)
barplot(rbind(colSums(norm(t(sfs.2d))),colSums(pop1.pop2)),be=T)
}
if(FALSE){
##just redo the angsd and optimization
##git checkout master ;make clean;make
system("../angsd -glf pop1.glf.gz -nind 12 -doSaf 1 -out pop1 -fai fai.fai -issim 1 -P 10")
system("../angsd -glf pop2.glf.gz -nind 8 -doSaf 1 -out pop2 -fai fai.fai -issim 1 -P 10")
system("../misc/realSFS pop1.saf 24 -P 60 >pop1.saf.idx.ml")
system("../misc/realSFS pop2.saf 16 -P 60 >pop2.saf.idx.ml")
system("../misc/realSFS 2dsfs pop1.saf pop2.saf 24 16 -P 60 >pop1.pop2.saf.idx.ml")
}
if(FALSE){
pop1 <- norm(exp(scan("pop1.saf.idx.ml"))[-1])
pop2 <- norm(exp(scan("pop2.saf.idx.ml"))[-1])
pop1.pop2 <- matrix(exp(scan("pop1.pop2.saf.idx.ml")),nPop1+1,byrow=T)
par(mfrow=c(1,2))
barplot(rbind(norm(table(p1.d)[-1]),pop1),be=T,main="varsites pop1")
barplot(rbind(norm(table(p2.d)[-1]),pop2),be=T,main="varsites pop2")
pop1.pop2[1,1] <- 0
pop1.pop2[nrow(pop1.pop2),ncol(pop1.pop2)] <- 0
pop1.pop2 <- norm(pop1.pop2)
range(norm(sfs.2d)-t(pop1.pop2))
## [1] -0.0005046856 0.0007019217
}
}