Main Page: Difference between revisions
Jump to navigation
Jump to search
(→Relate) |
|||
| Line 17: | Line 17: | ||
= [[Relate]]= | = [[Relate]]= | ||
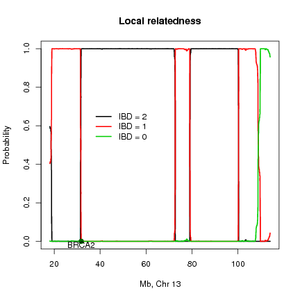
[[File:relate.png|thumb|Infered IBD sharing across a chromosome for a sib pair estimated using affy 500k data]] | |||
Revision as of 10:33, 15 August 2013
ANGSD
Analysis of Next Generation Sequencing Data
<classdiagram type="dir:LR"> [sequence data]->[genotype;likelihoods] [genotype;likelihoods]->[genotype;probabilities] [sequence files|bam files;SOAP files{bg:orange}]->[sequence data] [glf files|glfv3;soapSNP{bg:orange}]->[genotype;likelihoods] [genotype prob|beagle output{bg:orange}]->[genotype;probabilities] </classdiagram>
NgsAdmix
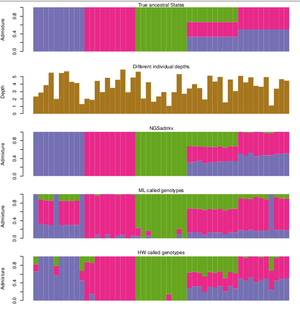
Infer the ancestry proportions from low depth NGS data. The principal is the same as other softwares such as FRAPPE and ADMIXTURE however, ngsAdmix also works when you have uncertainty in your data. This makes it ideal for medium and low depth sequencing data where many genotypes cannot be called without introducing errors or ascertainment bias.
Relate