RelateAdmix: Difference between revisions
| Line 65: | Line 65: | ||
</pre> | </pre> | ||
[[File:relAd.png]] | [[File:relAd.png|thumb]] | ||
NB!. Only use binary plink (.bed) since ADMIXTURE switches allele frequencies when using .ped files | NB!. Only use binary plink (.bed) since ADMIXTURE switches allele frequencies when using .ped files | ||
Revision as of 15:53, 21 August 2013
Brief description
This page contains information about the program called relateAdmix, which can be used to infer relatedness coefficients for pairs of individuals even if they are admixed. The program has both an R interface and a C interface. Below is a description of how to install and use each of them. To be able to infer the relatedness you will need to know the individuals admixture proportions and the allele frequencies in each of the possible populations. This can be done e.g. using the program Admixture as shown in the example of how to use the C interface.
Installation
Download
Installation of R package
wget http://www.popgen.dk/software/download/relateAdmix/relateAdmix_0.05.tar.gz R CMD INSTALL relateAdmix_0.05.tar.gz
Installation of C program
wget http://www.popgen.dk/software/download/relateAdmix/relateAdmix_0.05.tar.gz tar -xvzf relateAdmix_0.05.tar.gz cd relateAdmix/src/ mv CPP_Makefile Makefile make
Run example
Run example using R
After installing the package you can load it into R and try the example
library(relateAdmix) example(relate)
This shows an example of how to use the package. More information can be found in the man pages
?relate
Run example using C
After installing the program you can try running it on the example data set in the data folder, which consists of 50 individuals that are admixed from 2 source populations.
If you are in the src folder where you installed relateAdmix and you have the software Admixture installed this can be done as follows:
cd ../data
# First run Admixture using plink ".bed" to produce population specific allele frequencies (smallPlink.2.P)
# and individual ancestry proportions (smallPlink.2.Q).
# (note other programs can also be used, e.g. Structure and FRAPPE)
admixture smallPlink.bed 2
# Then run relateAdmix
../src/relateAdmix -plink smallPlink -f smallPlink.2.P -q smallPlink.2.Q -P 20
# plot the results in R (R needs to be installed)
Rscript -e "r<-read.table('output.k',head=T,as.is=T);pdf('rel.pdf');plot(r[,4],r[,5],ylab='k2',xlab='k1');dev.off()"
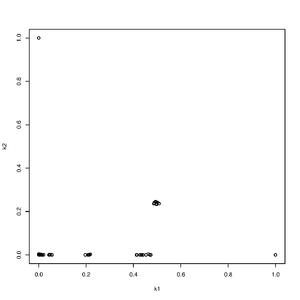
NB!. Only use binary plink (.bed) since ADMIXTURE switches allele frequencies when using .ped files
output file
example of output
ind1 ind2 k0 k1 k2 nIter 0 1 0.999941 0.000038 0.000021 26 0 2 0.999979 0.000010 0.000011 29 0 3 0.999953 0.000029 0.000018 26 0 4 0.999952 0.000023 0.000025 26 0 5 0.999972 0.000020 0.000007 26 0 6 0.999995 0.000003 0.000002 26 0 7 0.999995 0.000003 0.000002 26 0 8 0.999894 0.000069 0.000038 32 0 9 0.999894 0.000069 0.000038 32 0 10 0.999903 0.000071 0.000026 26 0 11 0.999903 0.000071 0.000026 26
The first two columns are the individuals number. The next three columns are the estimated relatedness coefficients and the last column is the number of iterations used
input files
example of the admixture proportion (for 3 populations)
0.531631 0.468359 0.000010 0.564461 0.435529 0.000010 0.850660 0.149330 0.000010 0.630527 0.369463 0.000010 0.747429 0.219346 0.033225 0.999980 0.000010 0.000010 0.999980 0.000010 0.000010 0.682072 0.317918 0.000010 0.000010 0.999980 0.000010 0.793133 0.206857 0.000010
echo row is an individual and each column is a population. The admxiture proportions for each individual must sum to 1
example of the allele frequency file (for 3 populations)
0.312722 0.208605 0.999990 0.881352 0.999990 0.966966 0.708206 0.838869 0.932119 0.427789 0.620694 0.532966 0.411998 0.622253 0.534072 0.427789 0.620694 0.532966 0.440817 0.581630 0.618751 0.733733 0.985281 0.953523 0.724083 0.451452 0.784607 0.811161 0.578612 0.787782
echo row is an SNP and each column is a population. When using plink file the allele frequency is the MAJOR allele frequency.