NgsRelate: Difference between revisions
| Line 3: | Line 3: | ||
=Installation= | =Installation= | ||
Primary repository is github. | |||
== Download Installation of C program == | |||
== Download | |||
<pre> | <pre> | ||
curl https://raw.githubusercontent.com/ANGSD/fastlate/master/fastlate.cpp >fastlate.cpp | |||
g++ fastlate.cpp -O3 -lz -o fastlate | |||
</pre> | </pre> | ||
Revision as of 20:18, 19 June 2015
Brief description
This page contains information about the program called NgsRelate, which can be used to infer relatedness coefficients for pairs of individuals for low coverage nags data by using genotype likelihoods. To be able to infer the relatedness you will need to know the population frequencies and have genotype likelihoods. This can be done e.g. using the program ANGSD as shown in the example.
Installation
Primary repository is github.
Download Installation of C program
curl https://raw.githubusercontent.com/ANGSD/fastlate/master/fastlate.cpp >fastlate.cpp g++ fastlate.cpp -O3 -lz -o fastlate
Run example
Run example using R
After installing the package you can load it into R and try the example
library(relateAdmix) example(relate)
This shows an example of how to use the package. More information can be found in the man pages
?relate
Run example using C
After installing the program you can try running it on the example data set in the data folder, which consists of 50 individuals that are admixed from 2 source populations.
If you are in the src folder where you installed relateAdmix and you have the software ADMIXTURE installed this can be done as follows:
cd ../data
# First run Admixture using a plink ".bed" as input to produce population specific allele
# frequencies (smallPlink.2.P) and individual ancestry proportions (smallPlink.2.Q).
# (note other programs can be used instead of Admixture, e.g. Structure and FRAPPE)
admixture smallPlink.bed 2
# Then run relateAdmix with plink bed, bim and fam files plus the Admixture output files as input
../src/relateAdmix -plink smallPlink -f smallPlink.2.P -q smallPlink.2.Q -P 20
# Plot the results in R (R needs to be installed)
Rscript -e "r<-read.table('output.k',head=T,as.is=T);pdf('rel.pdf');plot(r[,4],r[,5],ylab='k2',xlab='k1');dev.off()"
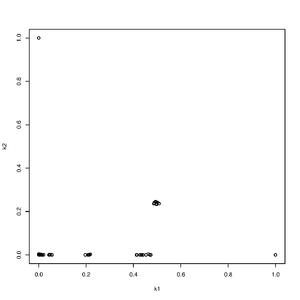
NB!. Only use binary plink (.bed) since ADMIXTURE switches allele frequencies when using .ped files
Output file format
Example of output
ind1 ind2 k0 k1 k2 nIter 0 1 0.999941 0.000038 0.000021 26 0 2 0.999979 0.000010 0.000011 29 0 3 0.999953 0.000029 0.000018 26 0 4 0.999952 0.000023 0.000025 26 0 5 0.999972 0.000020 0.000007 26 0 6 0.999995 0.000003 0.000002 26 0 7 0.999995 0.000003 0.000002 26 0 8 0.999894 0.000069 0.000038 32 0 9 0.999894 0.000069 0.000038 32 0 10 0.999903 0.000071 0.000026 26 0 11 0.999903 0.000071 0.000026 26
The first two columns are the individuals number. The next three columns are the estimated relatedness coefficients and the last column is the number of iterations used.
Input file format
The input consists of three files describignt the genotype data, a file with admixture proportions for each individual and a file with allele frequencies for each SNP for each source population. The genotype data files are plink bed/bim/fam files. And the remaining two files are in the output format for the program ADMIXTURE:
Example of the content of an admixture proportion file (for 3 populations)
0.531631 0.468359 0.000010 0.564461 0.435529 0.000010 0.850660 0.149330 0.000010 0.630527 0.369463 0.000010 0.747429 0.219346 0.033225 0.999980 0.000010 0.000010 0.999980 0.000010 0.000010 0.682072 0.317918 0.000010 0.000010 0.999980 0.000010 0.793133 0.206857 0.000010
Each row is an individual and each column is a population. The admixture proportions for each individual must sum to 1
Example of the allele frequency file (for 3 populations)
0.312722 0.208605 0.999990 0.881352 0.999990 0.966966 0.708206 0.838869 0.932119 0.427789 0.620694 0.532966 0.411998 0.622253 0.534072 0.427789 0.620694 0.532966 0.440817 0.581630 0.618751 0.733733 0.985281 0.953523 0.724083 0.451452 0.784607 0.811161 0.578612 0.787782
Each row is an SNP and each column is a population. When using plink files the allele frequency is the MAJOR allele frequency.
Citing and references
relateAdmix
Moltke, I, Albrechtsen, A (2013). RelateAdmix: a software tool for estimating relatedness between admixed individuals. Bioinformatics. pubmed bibtex
ADMIXTURE
D.H. Alexander, J. Novembre, and K. Lange. Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19:1655–1664, 2009.
change log
- 0.14 made more MAC usable (I think). Thanks to Paul Lott for reporting it and for suggestions and Thorfinn Sand for changing it
- 0.13 added extra check for file exists to give instant errors + changes all printf to fprintf(stderr,
- 0.11 changed threading to a fixed pool of threads
- 0.10 optimized code
- 0.09 added error for when the number of sites and individuals does not match between files
- 0.08 fixed a bug that would sometimes print an extra line when multiple threaded
- 0.07 fixed a small leak