ANGSD: Analysis of next generation Sequencing Data
Latest tar.gz version is (0.938/0.939 on github), see Change_log for changes, and download it here.
Glcomparison: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 63: | Line 63: | ||
aplot <- function(x,...){ | aplot <- function(x,...){ | ||
d<- | d<-(scan(x)) | ||
p <- 1-d[1]-d[length(d)] | p <- 1-d[1]-d[length(d)] | ||
barplot(norm(d[-c(1,length(d))]),col=2,legend=paste0("var=",round(p*100,3)),...) | barplot(norm(d[-c(1,length(d))]),col=2,legend=paste0("var=",round(p*100,3)),...) | ||
Latest revision as of 16:44, 5 February 2016
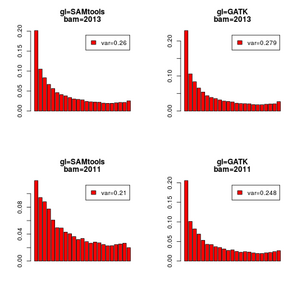
For some analysis the choice of GL model will make a huge impact, this is shown in the example where we estimate the sfs using -GL 1 and -GL 2, for two list of BAM files downloaded from the 1000genomes project website. First list are 12 CEU samples from 2011, and second list are the same 12 CEU samples but from 2013.
Below are the commands used
angsd -b new.list -minQ 20 -minMapQ 30 -isBed 1 -sites 1kg.accesible2.gz -GL 1 -out res/new.list.gl1 -doSaf 1 -r 1: -anc hg19ancNoChr.fa -P 6 angsd -b new.list -minQ 20 -minMapQ 30 -isBed 1 -sites 1kg.accesible2.gz -GL 2 -out res/new.list.gl2 -doSaf 1 -r 1: -anc hg19ancNoChr.fa -P 6 angsd -b old.list -minQ 20 -minMapQ 30 -isBed 1 -sites 1kg.accesible2.gz -GL 1 -out res/old.list.gl1 -doSaf 1 -r 1: -anc hg19ancNoChr.fa -P 6 angsd -b old.list -minQ 20 -minMapQ 30 -isBed 1 -sites 1kg.accesible2.gz -GL 2 -out res/old.list.gl2 -doSaf 1 -r 1: -anc hg19ancNoChr.fa -P 6
with
cat new.list NA06985.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam NA06994.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam NA07000.mapped.ILLUMINA.bwa.CEU.low_coverage.20130415.bam NA07056.mapped.ILLUMINA.bwa.CEU.low_coverage.20130415.bam NA07357.mapped.ILLUMINA.bwa.CEU.low_coverage.20130415.bam NA11829.mapped.ILLUMINA.bwa.CEU.low_coverage.20130415.bam NA11830.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam NA11831.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam NA11832.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam NA11840.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam NA11881.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam NA11992.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam
cat old.list NA06985.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA06994.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA07000.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA07056.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA07357.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA11829.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA11830.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA11831.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA11832.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA11840.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA11881.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam NA11992.mapped.ILLUMINA.bwa.CEU.low_coverage.20111114.bam
and 1kg.accesible2.gz are the accessible regions downloaded from the ucsc,and hg19ancNoChr.fa contains the ancestral state.
We do the optimisation with:
realSFS res/new.list.gl1.saf 36 -P 24 >res/new.list.gl1.saf.ml realSFS res/new.list.gl2.saf 36 -P 24 >res/new.list.gl2.saf.ml realSFS res/old.list.gl1.saf 36 -P 24 >res/old.list.gl1.saf.ml realSFS res/old.list.gl2.saf 36 -P 24 >res/old.list.gl2.saf.ml
and plot with
SAM <- system("ls res/*.ml|grep -v 2",intern=T)
GATK <- system("ls res/*.ml|grep 2",intern=T)
norm <- function(x) x/sum(x)
aplot <- function(x,...){
d<-(scan(x))
p <- 1-d[1]-d[length(d)]
barplot(norm(d[-c(1,length(d))]),col=2,legend=paste0("var=",round(p*100,3)),...)
}
png("glcomparison.png")
par(mfrow=c(2,2))
aplot(SAM[1],main=c("gl=SAMtools","bam=2013"))
aplot(GATK[1],main=c("gl=GATK","bam=2013"))
aplot(SAM[2],main=c("gl=SAMtools","bam=2011"))
aplot(GATK[2],main=c("gl=GATK","bam=2011"))
dev.off()