ANGSD: Analysis of next generation Sequencing Data
Latest tar.gz version is (0.938/0.939 on github), see Change_log for changes, and download it here.
PCA MDS
single read sampling approach for PCA or MDS
This function is new and works from version 0.912 and in the latest developmental version from github
Brief Overview
./angsd -doIBS -> angsd version: 0.911-26-gf1cb0e0-dirty (htslib: 1.3-1-gc72ae90) build(Apr 27 2016 11:15:33) -> Analysis helpbox/synopsis information: -> Command: ../angsd/angsd -doIBS -> Wed Apr 27 12:38:35 2016 -------------- abcIBS.cpp: -doIBS 0 (Sampling strategies) 0: no IBS 1: (Sample single base) 2: (Concensus base) -doCounts 0 Must choose -doCount 1 Optional -minMinor 0 Minimum observed minor alleles -minFreq 0.000 Minimum minor allele frequency -output01 0 output 0 and 1s instead of based -maxMis -1 Maximum missing bases (per site) -doMajorMinor 0 use input files or data to select major and minor alleles -makeMatrix 0 print out the ibs matrix -doCov 0 print out the cov matrix
Options
- -doIBS [int]
Print a single base from each individual at each position. 1: random sampled read. 2: Consensus base
- -doCounts [int]
Method requeres counting the different bases at each position. Therefore, -doCounts 1 must be used
- -doMajorMinor [int]
The covariance matrix can only be calculated for diallelic sites. Therefore, choose a methods for selecting the major and minor allele (see Inferring_Major_and_Minor_alleles). This can also be use if you only want to make this assumption for the IBS matrix or only want to print out bases that are either the major or minor.
- -minMinor [int]
Minimum observed minor alleles. The default in 0. If you do not use -doMajorMinor then the number of minor alleles are the sum of the 3 most uncommon alleles.
- --minFreq [float]
Minimum minor allele frequency. The default in 0. If you do not use -doMajorMinor then the frequency is the sum of the frequencies of the 3 most uncommon alleles.
- -output01 [int]
output the samples reads as 0 (for major) and 1s (for non major) instead of actual base
- -maxMis [int]
Maximum missing bases (per site) i.e. maximum number of uninformative individuals for the site
-makeMatrix [int] 1 prints out the pairwise IBS matrix. This is the avg. distance between pairs of individuals. Distance is zero if the base in the same and 1 otherwise.
- -doCov [int]
1 print out the covariance matrix.
Output
sampled bases *ibs.gz
This function will print the sampled based *ibs.gz.
Example of output *.ibs.gz with -doMajorMinor>0 and -output01 1
chr pos major minor ind0 ind1 ind2 ind3 ind4 ind5 ind6 ind7 1 14000873 A G 0 1 1 1 1 1 1 1 14001018 C T 0 1 1 1 1 1 1 1 14001867 G A 0 1 1 1 1 0 1 1 14002342 T C 1 1 1 1 1 -1 1 1 14002422 T A 0 1 1 1 1 0 -1 1 14003581 T C 0 1 1 1 1 1 1 1 14004623 C T 0 1 1 1 1 0 1 1 14006543 T G 0 -1 1 1 1 0 1 1 14007493 G A 0 0 1 -1 1 0 1 1 14007558 T C 0 0 1 1 -1 -1 1 1 14007649 A G 0 1 1 1 1 0 1 1 14008269 A G 1 1 0 -1 1 -1 1
Example of output *.ibs.gz with -doMajorMinor>0 and -output01 0
chr pos major minor ind0 ind1 ind2 ind3 ind4 ind5 ind6 ind7 1 13116 G T N G T T N G N T 1 13118 G A N G A A N G N A 1 14930 A G G G G A N N A N 1 15211 T G N G T G N N N G 1 54490 A G N G N G N N N N 1 54716 T C T C C C T N N N 1 58814 A G N G N G G G N N 1 62777 T A N N A N A A A N 1 63268 C T N T N T C N T N 1 63671 A G N G N N G G G N 1 69428 G T N G T N N T T N 1 69761 T A A A T A N A N N
Example of output *.ibs.gz with -doMajorMinor 0 and -output01 0
chr pos major ind0 ind1 ind2 ind3 ind4 ind5 ind6 ind7 ind8 1 13116 T N G T T N G N T T 1 13118 A N G A A N G N A A 1 14930 A G G G A N N A N G 1 15211 G N G T G N N N G G 1 54490 G N G N G N N N N A 1 54716 C T C C C T N N N C 1 58814 G N G N G G G N N G 1 62777 A N N A N A A A N A 1 63268 T N T N T C N T N N 1 63336 C C C C C C N C N N 1 63671 G N G N N G G G N N
chr is the chromosome
pos is the position major is the major allele
minor is the minor allele. Needs -doMajorMinor
indX is samples base for individual number X. if -output01 1 then it is 1 for major, 0 for non major and -1 for missing
sample based IBS matrix *.ibsMat
This function will print the pairwise IBS distance
Example of output *.ibsMat with -makeMatrix 1
0.000000 0.510638 0.606383 0.595745 0.545455 0.428571 0.510638 0.000000 0.154639 0.154639 0.108911 0.408602 0.606383 0.154639 0.000000 0.121212 0.137255 0.489362 0.595745 0.154639 0.121212 0.000000 0.106796 0.484211 0.545455 0.108911 0.137255 0.106796 0.000000 0.404040 0.428571 0.408602 0.489362 0.484211 0.404040 0.000000 0.577320 0.121212 0.181818 0.171717 0.097087 0.473684 0.536082 0.090000 0.138614 0.118812 0.047619 0.428571 0.262500 0.571429 0.702381 0.694118 0.632184 0.353659 0.458333 0.383838 0.484848 0.494949 0.398058 0.368421
Nind x Nind matrix with pairwise IBS distance
sample based covariance matrix *.covMat
This function will print the covariance matrix based on a single sampled read
Example of output *.covMat with -doCov 1
1.098251 -0.026225 -0.005617 -0.014726 -0.022438 -0.021786 -0.026225 1.115986 -0.017167 0.000735 -0.017163 -0.016899 -0.005617 -0.017167 1.074779 -0.015685 -0.019819 -0.015473 -0.014726 0.000735 -0.015685 1.072853 -0.013641 -0.007789 -0.022438 -0.017163 -0.019819 -0.013641 1.094612 -0.016045 -0.021786 -0.016899 -0.015473 -0.007789 -0.016045 1.059264 -0.005831 -0.009854 -0.001269 -0.002362 -0.018479 -0.011942 -0.015399 -0.020010 -0.001296 -0.022947 -0.006515 -0.003938 -0.001730 -0.040534 -0.002295 -0.017442 -0.024194 -0.007469 -0.016094 -0.015303 -0.018302 -0.022502 -0.030503 -0.001208 -0.122045 -0.106068 -0.103089 -0.104443 -0.110237 -0.103610 -0.106553 -0.100202 -0.104754 -0.109399 -0.107645 -0.111665 -0.108945 -0.102440 -0.105292 -0.101372 -0.107110 -0.106639
Nind x Nind covariance matrix
Model
IBS
pairwise distance between individuals where M in the number of sites with a read for both individuals. is the indicator function which is equal to one with the two individuals i and j have the same base and zero otherwise
Covariance
Allele frequency based on single reads.
where M in the number of sites with a read for both individuals. is 1 if individuals i for site m has the major allele and zero otherwise
MDS/PCA using R
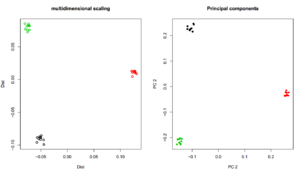
## MDS name <- "angsdput.ibsMat" m <- as.matrix(read.table(name)) mds <- cmdscale(as.dist(m)) plot(mds,lwd=2,ylab="Dist",xlab="Dist",main="multidimensional scaling",col=rep(1:3,each=10)) name <- "angsdput.covMat" m <- as.matrix(read.table(name)) e <- eigen(m) plot(e$vectors[,1:2],lwd=2,ylab="PC 2",xlab="PC 2",main="Principal components",col=rep(1:3,each=10),pch=16)
other fun stuff
## heatmap / clustering / trees name <- "angsdput.ibsMat" # or covMat m <- as.matrix(read.table(name)) #heat map heatmap(m) #neighbour joining plot(ape::nj(m)) plot(hclust(dist(m), "ave")




